Confined, we all follow the evolution of this damn virus. No ? What I am proposing to you through this article is to collect by yourself the official data from Public Health as well as from the Ministry of Health in order to do your own analyzes. These data are deposited in github: https://github.com/opencovid19-fr/data but they are in YAML format and require some alterations to be usable.
Index
Format de données YAML
YAML data format
This format is a fairly easy to use format, a kind of very simplified json that looks like structured data through indentation. Here is an example file:
date: "2020-01-24"
source:
nom: Ministère des Solidarités et de la Santé
url: https://solidarites-sante.gouv.fr/IMG/pdf/200124-cp_coronavirus.pdf
archive: https://web.archive.org/web/20200322183046/https://solidarites-sante.gouv.fr/IMG/pdf/200124-cp_coronavirus.pdf
donneesNationales:
casConfirmes: 3
hospitalises: 3
deces: 0
donneesRegionales:
- nom: Île-de-France
code: REG-11
casConfirmes: 2
hospitalises: 2
- nom: Nouvelle-Aquitaine
code: REG-75
casConfirmes: 1
hospitalises: 1Fortunately, and as often Python provides a library to handle this type of format automatically and convert it into a usable format simply: JSON.
To do this, you must install the package:
pip install yaml
Then you can read the file with the method:
<pre class="wp-block-syntaxhighlighter-code">json_data = yaml.load(<Contenu du fichier>, Loader=yaml.FullLoader)</pre>
The value of json_data is returned in JSON format.
Raw data recovery
We will retrieve the data directly by querying Github. To do this, you must query Github directly on the data (raw) in: https://raw.githubusercontent.com/opencovid19-fr/data/master/sante-publique-france/
Each file is in YYYY-MM-DD.yaml format so it will be necessary to scan all the dates from the beginning to today.
r = requests.get('https://raw.githubusercontent.com/opencovid19-fr/data/master/sante-publique-france/2020-03-24.yaml')
if (r.status_code==200):
print (yaml.load(r.text, Loader=yaml.FullLoader))
Creating the file name to recover is not very complex, you just have to recover all the data one after the other and recompose the file name in GitHub:
hier = date.today() + timedelta(-1)
shier = hier.strftime("%Y-%m-%d")
fichier = 'https://raw.githubusercontent.com/opencovid19-fr/data/master/sante-publique-france/' + shier + '.yaml'
If we want to recover yesterday’s file, here is a small function that will do it in a few lines:
def RecupCovid19Hier():
hier = date.today() + timedelta(-1)
shier = hier.strftime("%Y-%m-%d")
fichier = 'https://raw.githubusercontent.com/opencovid19-fr/data/master/sante-publique-france/' + shier + '.yaml'
out = ""
r = requests.get(fichier)
if (r.status_code==200):
out = yaml.load(r.text, Loader=yaml.FullLoader)
return out
Let’s retrieve the data from San Santé Publique France
The data was released as of January 23, 2020.
Here is the function that will allow you to recover data through YAML files (one per day) using the principles explained above.
import yaml
import requests
import time
from datetime import datetime, timedelta, date
import pandas as pd
# INITIALISATION DE VARIABLES GLOBALES
sDateDebut = "2020-01-23"
sAujourdhui = date.today().strftime("%Y-%m-%d")
dDemarrageDate = datetime.fromisoformat(sDateDebut)
cols = ['Date',
'FR Tot Cas Confirmés',
'FR Tot Décès',
'WW Tot Cas Confirmés',
'WW Tot Décès',
'Source',
'FR Tot Guéris',
'FR Tot Hospitalisés'
]
# FONCTIONS
def recupDataWithException2(_jsonout, _index1, _index2):
try:
data = _jsonout[_index1][_index2]
except:
data = 0
return data
def AjouteSantePubliqueFrLigne(jsonout, df):
df = df.append({
cols[0] : jsonout['date'] ,
cols[1] : recupDataWithException2(jsonout, 'donneesNationales', 'casConfirmes') ,
cols[2] : recupDataWithException2(jsonout, 'donneesNationales', 'deces'),
cols[3] : recupDataWithException2(jsonout,'donneesMondiales', 'casConfirmes'),
cols[4] : recupDataWithException2(jsonout,'donneesMondiales', 'deces'),
cols[5] : 'sante-publique-france',
cols[6] : 0,
cols[7] : 0
} ,
ignore_index=True)
return df
def ajouteLigneVide(df, madate):
df = df.append({
cols[0] : madate ,
cols[1] : 0,
cols[2] : 0,
cols[3] : 0,
cols[4] : 0,
cols[5] : 'no-data',
cols[6] : 0,
cols[7] : 0
} ,
ignore_index=True)
return df
# RECUPERATION DONNEES SANTE PUBLIQUE FRANCE
def RecupereDonneesSantePubliqueFrance():
print ('--> Démarrage du process')
sDateParcours = sDateDebut
i=1
requesReturnCode=200
yamls = pd.DataFrame()
while (sDateParcours != sAujourdhui):
myDate = dDemarrageDate + timedelta(days=i)
sDateParcours = myDate.strftime("%Y-%m-%d")
fichier = 'https://raw.githubusercontent.com/opencovid19-fr/data/master/sante-publique-france/' + sDateParcours + '.yaml'
#print ('--> sante-publique-france / ', sDateParcours)
req = requests.get(fichier)
requesReturnCode = req.status_code
if (requesReturnCode==200):
yamlout = yaml.load(req.text, Loader=yaml.FullLoader)
yamls = AjouteSantePubliqueFrLigne(yamlout, yamls)
else:
print ("(*) Pas de données pour ", myDate.strftime("%Y-%m-%d"))
# ajoute une ligne vide
yamls = ajouteLigneVide(yamls, myDate.strftime("%Y-%m-%d"))
i=i+1
print ('--> Fin du process')
return yamls
data = RecupereDonneesSantePubliqueFrance()
data
This function therefore returns a Pandas dataframe which contains Public Health France data.
Date FR Tot Cas Confirmés FR Tot Décès FR Tot Guéris FR Tot Hospitalisés Source WW Tot Cas Confirmés WW Tot Décès
0 2020-01-24 3.0 0.0 0.0 0.0 sante-publique-france 897.0 26.0
1 2020-01-25 3.0 0.0 0.0 0.0 sante-publique-france 1329.0 41.0
2 2020-01-26 3.0 0.0 0.0 0.0 sante-publique-france 2026.0 56.0
3 2020-01-27 3.0 0.0 0.0 0.0 sante-publique-france 2820.0 81.0
4 2020-01-28 4.0 0.0 0.0 0.0 sante-publique-france 0.0 0.0
... ... ... ... ... ... ... ... ...
64 2020-03-28 37575.0 2314.0 0.0 0.0 sante-publique-france 591971.0 27090.0
65 2020-03-29 0.0 0.0 0.0 0.0 no-data 0.0 0.0
66 2020-03-30 0.0 0.0 0.0 0.0 no-data 0.0 0.0
67 2020-03-31 0.0 0.0 0.0 0.0 no-data 0.0 0.0
68 2020-04-01 0.0 0.0 0.0 0.0 no-data 0.0 0.0
69 rows × 8 columnsData recovery from the Ministry of Health
The recovery will follow the same principle. However, here we are not going to create a new Pandas dataframe , we will rather enrich the data already retrieved previously with those of the ministry (in particular we recover some additional information such as the number of cured, the number of hospitalizations).
Here is the function that retrieves and enriches the previous dataframe:
# RECUPERATION DONNEES SANTE PUBLIQUE FRANCE
def RecupereDonneesMinistereSante(yamls):
i=1
sDateParcours = sDateDebut
print ('--> Démarrage du process')
while (sDateParcours != sAujourdhui):
myDate = dDemarrageDate + timedelta(days=i)
sDateParcours = myDate.strftime("%Y-%m-%d")
#print("Traintement date: ", dateParcours)
fichier = 'https://raw.githubusercontent.com/opencovid19-fr/data/master/ministere-sante/' + sDateParcours + '.yaml'
req = requests.get(fichier)
requesReturnCode = req.status_code
if (requesReturnCode==200):
yamlout = yaml.load(req.text, Loader=yaml.FullLoader)
if (yamls[cols[5]][i-1] == "no-data"):
print ("(*) Modifie les données manquantes pour la date du ", myDate.strftime("%Y-%m-%d"))
# Modifie toute la ligne
yamls.loc[i-1, cols[1]] = recupDataWithException2(yamlout, 'donneesNationales', 'casConfirmes')
yamls.loc[i-1, cols[2]] = recupDataWithException2(yamlout, 'donneesNationales', 'deces')
yamls.loc[i-1, cols[3]] = recupDataWithException2(yamlout, 'donneesMondiales', 'casConfirmes')
yamls.loc[i-1, cols[4]] = recupDataWithException2(yamlout, 'donneesMondiales', 'deces')
yamls.loc[i-1, cols[5]] = "ministere-sante"
# Ajoute les nouveaux champs
yamls.loc[i-1, cols[6]] = recupDataWithException2(yamlout, 'donneesNationales', 'hospitalises')
yamls.loc[i-1, cols[7]] = recupDataWithException2(yamlout, 'donneesNationales', 'gueris')
i=i+1
print ('--> Fin du process')
return yamls
Some finishing touches on the data
The raw data retrieved is cumulative data. In addition, we note that some days were not officially published (no-data). The consequence is that the data present “holes” which is a bit annoying for cumulative data. We will fill these holes:
def Bouchetrous(yamls):
# On reparcourre toutes les lignes déjà récupérées précédemment
i=1
for index, row in yamls.iterrows():
if (yamls[cols[5]][i-1] == "no-data"):
for j in [1, 2, 3, 4, 6, 7]:
yamls.loc[i-1, cols[j]] = yamls[cols[j]][i-2]
#yamls[cols[j]][i-1] = yamls[cols[j]][i-2]
i += 1
return yamls
For those who want to go further, it might also be interesting to “disaggregate” the data, or do other operations.
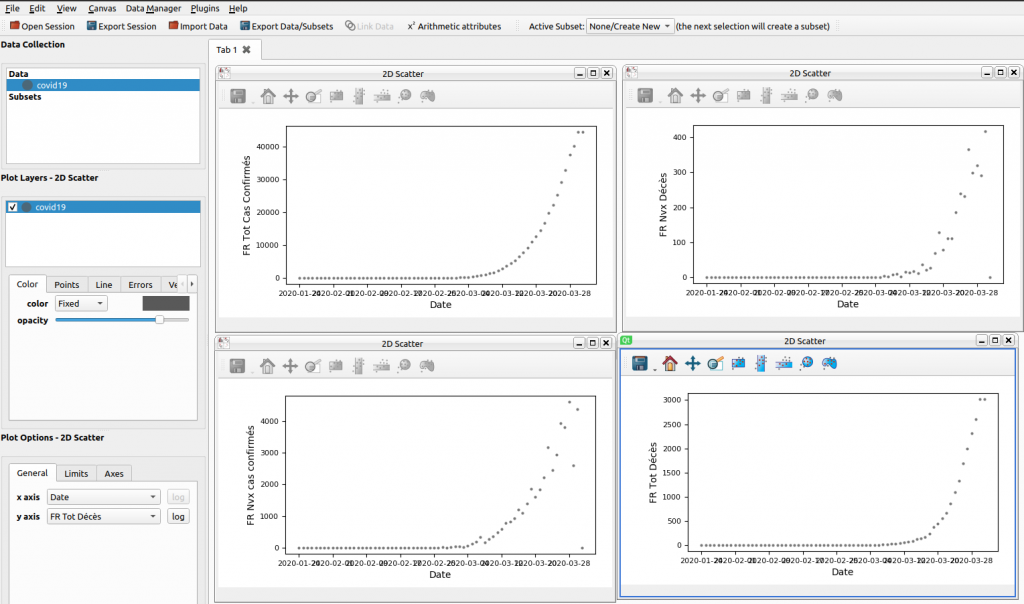
The result ?
Here is an overview of the data, which unfortunately shows us that the peak of the epidemic has not yet been reached.
You can grab the full code from GitHub